Abstract
The giant panda (Ailuropioda melanoleuca), a unique relic species in China and a global biodiversity conservation symbol, faces the threat of canine distemper virus (CDV). Vaccinating domestic dogs in panda habitats against CDV is crucial, yet the associated risks remain understudied. We investigated the safety of CDV vaccination in 69 domestic dogs within panda habitats, employing enzyme-linked immunosorbent assay for CDV antibodies and quantitative reverse transcription polymerase chain reaction for viral RNA, a marker of viral shedding. Results revealed that vaccinated dogs posed a risk through viral shedding, mainly starting from the ninth day post-vaccination. Unvaccinated dogs exhibited increased CDV antibodies and subsequent shedding, with phylogenetic analysis confirming infection from vaccinated dogs in shared kennels. Dogs with higher initial antibodies displayed reduced shedding and a markedly abbreviated shedding duration (6.7 days) than those with lower initial antibodies (9.8 days). Habitat area analysis revealed substantial overlaps between domestic dogs and wild giant pandas in two nature reserves in China. To safeguard wildlife, particularly giant pandas, we recommend restricting vaccinated dogs’ activity for at least three weeks post-vaccination, complementing existing management practices. We advocate collaborative efforts among local authorities, reserve management and villagers for effective vaccination and post-vaccination management of domestic dogs.
Introduction
The global environment is undergoing change, and animal infectious diseases are becoming one of the major threats to the survival of wild animals, presenting a significant challenge to the conservation of global biodiversity. The giant panda (Ailuropioda melanoleuca) is a unique relic species in China and also a flagship species of biodiversity conservation worldwide1. As indicated by the findings of the Fourth National Survey on Giant Pandas, the current population of wild giant pandas is estimated to be 1864 individuals2. The distribution of wild giant pandas is confined to six mountain ranges: Qinling, Minshan, Qionglai, Daxiangling, Xiaoxiangling, and Liangshan Mountains. The population of these wild giant pandas is fragmented into 33 distinct and isolated populations with limited inter-population connectivity3. In recent years, a decline in the number of giant panda populations and a degradation in their habitat quality has been observed. This trend has been attributed to both natural disasters and human activities. Moreover, periodic outbreaks of epidemic diseases such as canine parvovirus and rotavirus in the giant panda habitat pose a significant threat to the survival of the species and attract increasing attention from researchers and conservationists alike4,5.
Canine distemper (CD), caused by canine distemper virus (CDV) belonging to the genus Morbillivirus of the family Paramyxoviridae, is a highly contagious and fatal disease6. It has been reported to infect giant pandas in China since 1994 and resulted in the deaths of several captive individuals7,8,9. The CDV outbreak in 2015 shows that infected pandas were difficult to cure, even with prompt treatment. This high rate of infection and mortality has the potential to threaten the survival of wild giant pandas and other rare species in their habitat10.
To prevent CD in giant pandas, vaccination is crucial11. However, there is currently no CDV vaccine specifically designed for giant pandas, and vaccines designed for carnivores such as dogs have been shown to be insufficient in establishing long-term protection12,13. Furthermore, the presence of thousands of domestic dogs in panda habitats increases the risk of CDV transmission to wildlife, including giant pandas8. In a CDV outbreak in China, the strains isolated from infected giant pandas show high homology with strains from raccoon dogs and domestic dogs14. Large-scale vaccination programs for domestic dog reservoirs have been implemented in some national parks, such as the Serengeti National Park and the Bale Mountains National Park, as the main approach to protect endangered wildlife15,16. Similarly, vaccination programs for domestic dogs are underway in some core areas of giant panda habitats in an orderly manner. Despite the increasing recognition of the importance of vaccination in wild giant panda conservation, post-vaccination management often lacks adequate attention. Most commercial core vaccines for dogs, containing CDV strains, are live attenuated vaccines, and the profile of viral shedding after vaccination can pose a potential threat to giant pandas and other sensitive wild species in their habitats. However, there is a lack of evidence-based guidance on the subsequent management of vaccinated dogs living in giant panda habitats.
In this study, enzyme-linked immunosorbent assay (ELISA) was used to detect changes in CDV antibodies in dogs before and after vaccination, and quantitative reverse transcription polymerase chain reaction (qRT-PCR) was used to detect viral RNA in the feces, urine, and secretions of dogs. This study aims to explore the pattern of viral shedding after vaccination and to provide scientific evidence for improving the precise prevention and conservation management of epidemic diseases in giant pandas and sympatric wild animals.
Results
Detection of CDV antibody by ELISA (IgG)
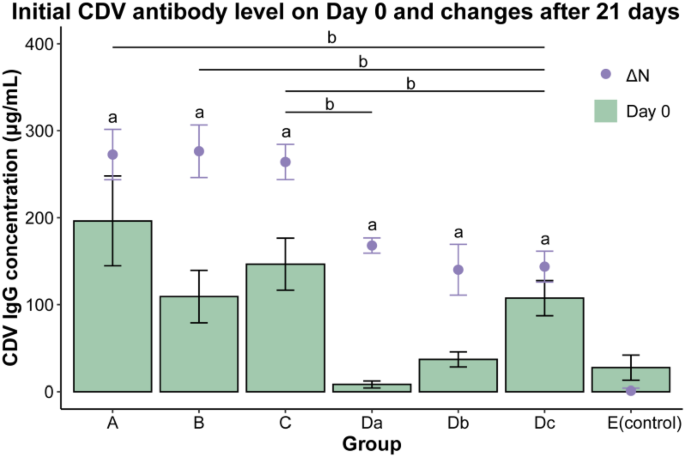
The serum CDV antibody concentrations of dogs on Day 0 and Day 21 were measured, and the increases in CDV antibody levels were calculated over a period of 21 days after vaccination (Fig. 1). The cut-off value for the ELISA was 0.158. Statistical analysis was performed using the K-S test and one-way ANOVA. The control group, Group E, showed no significant increase in CDV IgG levels. On the other hand, the three vaccine groups (groups A, B and C) displayed significant increases in serum CDV IgG levels 21 days after vaccination, in comparison to group E (p < 0.05). No significant differences in antibody level increases were observed among these three vaccine groups (p > 0.05), indicating that there was no notable variation in the immune response between three vaccines. Additionally, significant increases in serum CDV IgG levels were found in all three sub-groups of group D (Da, Db, and Dc) (p < 0.05), with no significant difference in the increases of serum CDV IgG levels among them (p > 0.05).
Initial CDV antibody level and changes in antibody level of dogs 21 days after vaccination. ΔN refers to the change in IgG concentration on Day 0 and Day 21 (IgG concentration on Day 21 minus IgG concentration on Day 0). Da, dogs in this sub-group lived with a dog from group A; Db, dogs in this sub-group lived with a dog from group B; Dc, dogs in this sub-group lived with a dog from group C; E, the control group, dogs in this group were not vaccinated and housed separately. No significant differences were observed among ΔN of three vaccine groups and ΔN of sub-groups (p > 0.05). The notation “a” indicates a significant difference in ΔN when compared to the control group (p < 0.05). Specifically, the notation "b" denotes a significant difference in ΔN between the two connected groups (p < 0.05). The ΔN of group Da showed a significant difference compared to group C (p < 0.05). The ΔN of group Dc showed significant differences when compared to groups A, B, and C (p < 0.05).
Detection of CDV by qRT-PCR
We conducted daily detection of ocular and nasal secretions, as well as feces and urine samples, using qRT-PCR. The standard curve was generated using linear regression based on standards and the virus copies in samples were calculated using the linear regression equation as follows: y=-3.232x + 42.841, with a correlation coefficient of R2 = 0.992.
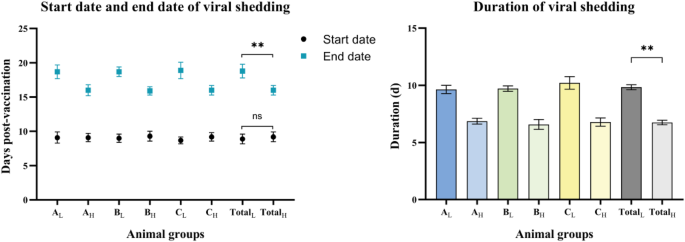
The incidence of positive results in ocular and nasal secretions was documented for each group post-vaccination (Appendix C). No evidence of viral shedding was observed in the ocular and nasal secretions of the dogs in the study from Day 0 to Day 7. In the three vaccine groups, 10–30% of dogs were detected positive from Day 8, and 100% of dogs in these groups were detected positive from Day 10 to Day 15. After this time, the detection of viral shedding in vaccinated dogs gradually decreased, with no dogs being detected positive on Day 21. It was noted that, although the dogs in group D were not vaccinated, samples taken from these dogs tested positive and their viral shedding started on Day 15 and ended on Day 20. Dogs in group E were consistently detected as negative during the study. The start and end dates of viral shedding in ocular and nasal secretions in three vaccine groups were shown (Fig. 2). The three vaccine groups were further divided into dogs with low antibody levels and dogs with high antibody levels. In addition, dogs with low antibody levels and dogs with high antibody levels were analyzed as a whole, respectively, as shown in group Total. Generally, viral shedding started on the ninth day after vaccination. The mean duration of viral shedding in dogs with low initial CDV antibody levels was 9.8 days (SD 1.2), significantly longer than the mean 6.7 days (SD 0.9) in dogs with high initial CDV antibody levels (p < 0.01). Compared with dogs with low initial CDV antibody levels, dogs with high CDV antibody levels had a later appearance and earlier disappearance of viral shedding, with a 32.3% shorter duration.
The start dates, end dates and duration of viral shedding in ocular and nasal secretions in three vaccine groups. The three vaccine groups were further divided into dogs with low antibody levels and dogs with high antibody levels, as shown in the figure (L and H). In addition, dogs with low antibody levels and dogs with high antibody levels were analyzed as a whole, respectively, as shown in group Total. The duration of viral shedding in dogs with low initial CDV antibody levels was 9.8 days (SD 1.2), significantly longer than the mean 6.7 days (SD 0.9) in dogs with high initial CDV antibody levels (p < 0.01).
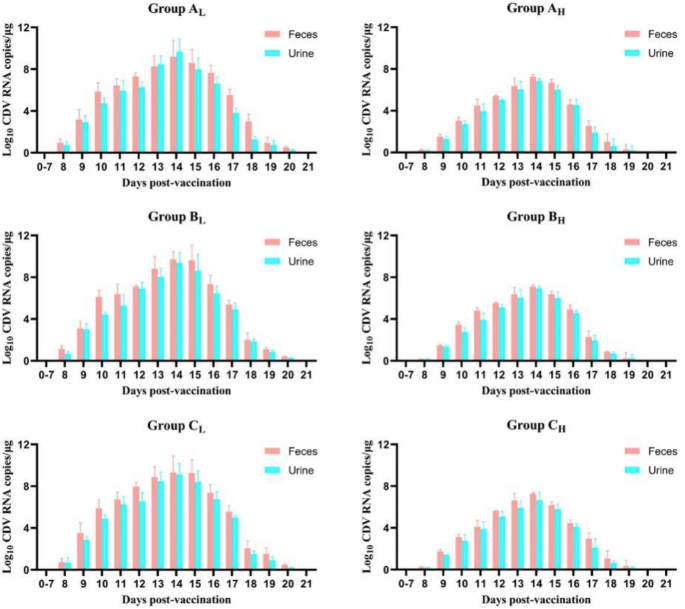
Regarding feces and urine samples, we observed fluctuations in viral load among the three vaccine groups (Fig. 3). The viral shedding curves of dogs after vaccination with one of three commercial vaccines were close to normal distribution. We detected positive for viral shedding from Day 8 to Day 20. No significant difference was observed in viral shedding in the three low antibody level groups (p > 0.05), nor in three high antibody level groups (p > 0.05). Notablely, viral load in feces and urine samples of dogs with low antibody levels was significantly higher than in dogs with high antibody levels in all three vaccine groups (p < 0.01). Both the low antibody level and high antibody level groups demonstrated the greatest extent of viral shedding on Day 14. The high antibody level groups exhibited significantly lower viral loads in both feces and urine samples, with a maximum of 1.57 × 107 copies/µL compared with 1.58 × 109 copies/µL in the low antibody level groups (p < 0.01). On Day 20, the viral shedding of dogs with high antibody levels was virtually undetectable. Subsequently, all feces and urine samples tested negative after Day 21.
Viral shedding in feces and urine samples after vaccination in vaccine groups. It shows the viral shedding dynamics of domestic dogs vaccinated with 3 commercial vaccines in 21 days. We detected positive for viral shedding from Day 8 to Day 20. No significant difference was observed in viral shedding in the three low antibody level groups (p > 0.05), nor in the high antibody level groups (p > 0.05). However, regardless of duration time and total amount, the viral shedding of dogs with low antibody levels was significantly more intense than in dogs with high antibody levels.
Phylogenetic analysis
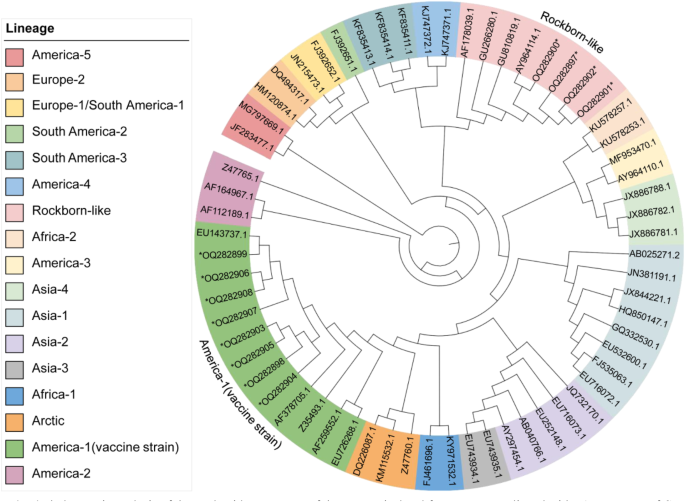
The H gene sequences of virus isolates in group D were amplified and sequenced through RT-PCR. The nucleotide sequence analysis of the H gene showed that the isolates from group Da were highly identical to the Vanguard vaccine that was used in group A, their co-living group (with 99.61%/ 99.55%/ 99.55% identity). The three isolates from group Db were found to be 99.74%, 99.67% and 99.67% identical to NOBIVAC, the vaccine used in group B. Similarly, the three isolates from group Dc showed 99.55%, 99.41% and 99.67% identity with EURICAN, the vaccine used in group C. The phylogenetic analysis, based on the H gene, demonstrated that these isolates belonged to the America-1 and Rockborn-like clusters, displaying homology with their respective vaccine strains and distinct from the wild strains prevalent in Asia, which belonged to the Asia-1 or Asia-2 cluster (Fig. 4).
Phylogenetic analysis of the nucleotide sequences of the H gene isolated from group D, aligned with 51 sequences of CDV reference strains in GenBank. The information for each reference strain is shown in Appendix D. Phylogenetic relationships were inferred using the Maximum Likelihood method and the Tamura-Nei model, generated with 1000 bootstrap replicates. The 12 sequences obtained in this study are marked with an asterisk and belong to the America-1 and Rockborn-like lineages, which are also highlighted in the figure (GenBank accession numbers: Vanguard-OQ282897, NOBIVAC-OQ282898, EURICAN-OQ282899, Dog D1 to Dog D9-OQ282900 to OQ282908).
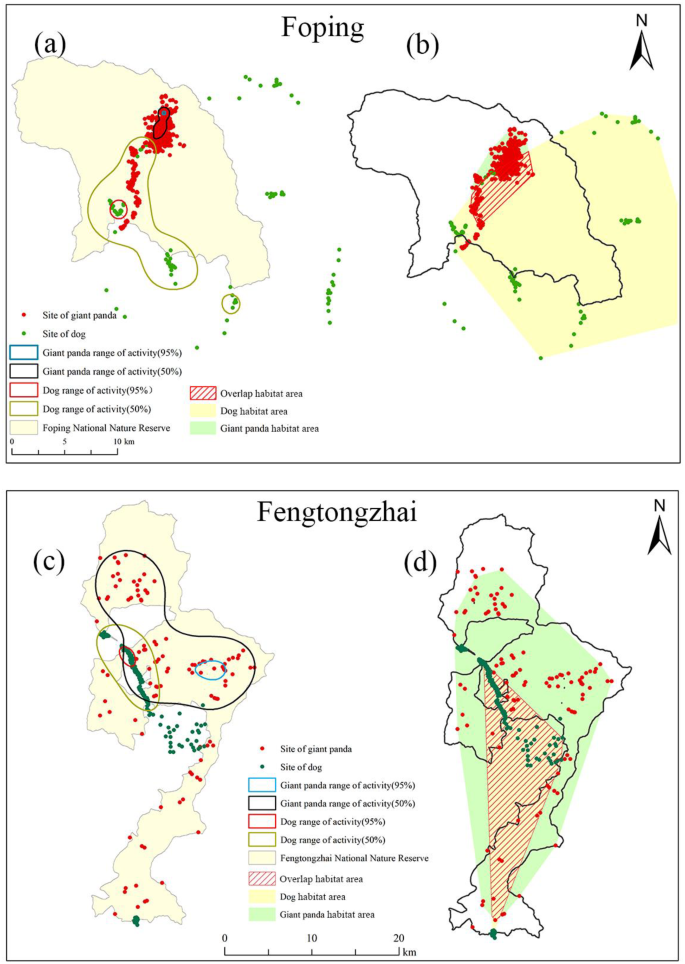
Habitat area overlap analysis between giant pandas and domestic dogs
The habitat area overlap analysis was performed on a total of 12 wild giant pandas (Foping, n = 6; Ya’an, n = 6) and 20 domestic dogs (Foping, n = 8; Ya’an, n = 12) (Fig. 5). In Foping, the results showed no core area overlap between pandas and dogs in 95% and 50% fixed kernel estimator (FKE). The minimum convex polygon (MCP) analysis indicated that the combined size of the habitat area of dogs tracked was 378.5 km2, while that of giant pandas was 30.7 km2. The MCP method revealed a substantial habitat area overlap of 61.3% between pandas and dogs in the vicinity of the FPNR. In Fengtongzhai Nature Reserve (FTNR), FKE showed a clear core area overlap between wild pandas and domestic dogs. The MCP analysis further supported this result, with a high habitat area overlap of 40.0% between wild pandas (455.2 km2) and domestic dogs (146.2 km2) in FTNR.
Habitat area analysis of wild giant pandas and domestic dogs around the Foping Nature Reserve (FPNR) and Fengtongzhai Nature Reserve (FTNR). Fixed kernel estimator (FKE) analysis of core areas was conducted for both wild pandas and domestic dogs in the vicinity of FPNR (a) and FTNR (c). The results of the Minimum Convex Polygon (MCP) analysis of habitat areas and overlap areas between domestic dogs and wild pandas around FPNR (b) and FTNR (d) are also shown. Map generated using ArcGIS v10.5 (Environmental Systems Research Institute Inc., Redlands, USA), with map data sourced from Foping National Nature Reserve and Fengtongzhai National Nature Reserve.
Discussion
Canine distemper virus has been shown to pose a significant threat to a diverse range of species, including carnivores such as Ailuridae, Mustelidae, Felidae, Phocidae, and non-human primates like Macaca17,18,19,20,21. This highly contagious and often fatal disease has become a major issue in global wildlife conservation, as evidenced by its impact on several species worldwide22. One prominent example of this impact can be seen in the Serengeti National Park in Tanzania, where a CDV-related morbillivirus outbreak caused a decline of approximately one-third in the population of lions (Panthera leo)23. In the United States, a similar outbreak of CDV almost led to the extinction of the black-footed ferret (Mustela nigripes)24. In the case of giant pandas, a CDV outbreak resulted in the death of five captive individuals, who presented with clinical symptoms related to CDV infection9. Given the low population density and fragmented distribution of wild giant pandas, the potential risk of CDV on this species has raised serious concerns for its survival and conservation.
In fact, CDV has been close to wild giant pandas. A high seroprevalence (89.9%) of CDV antibodies was detected among the 69 dogs in the vicinity of the FTNR, based on the initial antibody levels. This indicated that animals in this area, including these dogs that had no history of receiving canine core vaccines, were at high risk of exposure to CDV. This observation was consistent with previous studies in unvaccinated dog populations living near wildlife habitats. For instance, a study in Uganda found a 100% prevalence of CDV antibodies in 184 dog samples from three different national parks25. Additionally, a study in Bolivia recorded a high seroprevalence of 93% in 40 domestic dogs living near a national park26. According to data from national rabies surveillance sites in 2020, the average immunization rate for rabies vaccines in dogs in China is only around 30%27. This rate may be even lower in rural areas28. Given that rabies vaccination is a mandatory measure in China, such a low vaccination rate suggests that the immunization rate for the CDV vaccine may be even less optimistic. In the vaccine experiment of this study, none of the 69 dogs surveyed had a history of vaccination. Another survey conducted in villages surrounding a giant panda reserve also indicated a high proportion of free-roaming dogs and extremely low vaccination rates29. It suggested possible CDV circulations around giant panda reserves, which had not received adequate attention previously. Local authorities and natural reserve management departments should establish a regular vaccination and supervision system for domestic dogs within nature reserves, and provide necessary resources and support. Communities should actively organize villagers to support domestic dog vaccination programs, and supervise and manage the feeding and vaccination status of domestic dogs. We strongly encourage educational training for residents in reserves to raise awareness of vaccinating their dogs. Additionally, we are collaborating with reserve management departments to implement free commercial core vaccinations for dogs around the reserves.
A significant population of domestic dogs, utilized as guards, are kept in villages near FTNR and FPNR with a low vaccination rate. Our habitat area overlap analysis revealed that these dogs were capable of invading the habitats of giant pandas at any moment for food and water, with a high degree of overlap observed both in FTNR (40.0%) and FPNR (61.3%) (Fig. 5). Although 95% FKE and 50% FKE showed no overlap in FPNR, it was observed that the tracked giant panda had traveled through the core habitat area of domestic dogs. Previous research also demonstrated significant habitat overlap between domestic dogs and wild giant pandas in FPNR during cold seasons10. In FTNR, the habitat area overlap between giant pandas and domestic dogs was even greater, indicating an increased likelihood of direct or indirect contact between the two species and a higher risk of disease transmission. The likelihood of CDV transmission is heightened as dogs and pandas may share the same habitat or drink from the same water sources.
Domestic dogs have been widely documented as the source of canine distemper virus infections in wildlife30,31. The full genome of the strain isolated from a giant panda that died from canine distemper showed the highest nucleotide homology with the CDV PS strain isolated from a dog9. However, the application of CDV vaccination to dogs in giant panda habitats may not always result in desired conservation outcomes. Small carnivores such as foxes, minks, and raccoon dogs can act as reservoirs, facilitating virus transmission between dogs and giant pandas. Intermediate hosts increase cross-species transmission likelihood and may drive viral adaptation and evolution, enhancing pathogenicity and transmissibility32. In some cases, attenuated live vaccine strains can revert to virulence, especially in immunocompromised animals, making vaccinated dogs new transmission sources. Additionally, interactions between vaccine and wild-type strains may lead to new viral variants, increasing the risk of vaccinated dogs spreading the virus33. Although commercial core vaccines are deemed safe for dogs, there remains a limited understanding of the potential effects of these vaccine strains on giant pandas. Previous studies have reported incidents of vaccine-induced infections in red pandas34, as well as frequent vaccine-induced infections in ferrets and minks35,36. Furthermore, as most commercial CDV vaccines are live attenuated vaccines, the potential for viral shedding in vaccinated dogs can present a significant threat to giant pandas. The varying sensitivity of different animals to CDV vaccine strains raises concerns regarding vaccine-induced infections in giant pandas. CDV vaccine strains and wild strains carried by dogs have the potential to cross species, thus posing a threat to wildlife such as giant pandas14,30. To better guide herd immunity management in domestic dogs and reduce the threat to giant pandas and the associated species within their habitats, it is crucial to thoroughly understand the implications of viral shedding in vaccinated dogs and the associated risks to giant pandas.
In this study, all three vaccine groups (A, B, and C) demonstrated evidence of viral shedding, confirming our concerns about the potential transmission of the virus from vaccinated dogs (Fig. 3). The ocular and nasal secretions data were presented as positivity rates rather than quantified viral load due to methodological considerations (Appendix C). Specifically, the viral loads in these types of secretions were highly variable. In contrast, the feces and urine samples consistently provided quantifiable levels of viral load, allowing for a more detailed analysis. Additionally, we observed significant increases in CDV antibody levels in group D, which consisted of unvaccinated dogs. Furthermore, feces and urine samples collected from these dogs tested positive for CDV from Day 16 to Day 17. To clarify the source of CDV detected in group D, we sequenced and phylogenetically analyzed nine isolates from groups Da, Db, and Dc. Because of the highest variability, the H gene has been widely used in genetic and phylogenetic analysis37,38. The analysis of the H gene showed that the isolates from group Da were most closely related to Vanguard, the vaccine strain used in group A and belonged to the same cluster as Vanguard. Similarly, the isolates from groups Db and Dc were also closely related to their respective vaccines (Fig. 4). It should be clarified that despite data from phylogenetic analyses showing a very close nucleotide proximity between sequences detected in vaccinated dogs and vaccine sequences, it is still possible that the strains detected in the study were not vaccine strains. In addition, it is noteworthy that while the NOBIVAC and EURICAN vaccines belonged to the America-1 group, which comprises virus strains commonly used in current vaccines, the Vanguard vaccine was found to be positioned in Rockborn-like lineage, distinct from other commercial vaccine strains. Similar results were reported in several previous studies, where phylogenetic analysis of the H gene showed that Vanguard strain was closest to the wild strains from canine and lesser panda, respectively, and restriction fragment length polymorphism (RFLP) tests similarly showed that it reacted as a wild strain39,40. Certainly this vaccine strain underwent all the necessary tests to prove it was safe and effective for use in animals before being licensed. However, the susceptibility of other canids and wildlife to the Rockborn-like strain remains a concern. Moreover, the potential for the Rockborn-like lineage to interact with wild strains in the environment and regain virulence warrants further investigation.
In this study, unvaccinated dogs in group D began shedding the virus through ocular and nasal secretions starting on day 15 (Appendix C). Additionally, a rise in CDV antibody levels was detected, indicating an immune response to viral exposure. These results provide evidence of pathogen transmission during the viral shedding process of vaccinated dogs, raising concerns about the potential for secondary transmission to unvaccinated animals or wildlife. However, the presence of CDV RNA and antibodies does not confirm active infection or the ability to transmit the virus to other animals. This underscores the need for further research to directly confirm viral viability and infectious potential of the shed virus.
It is important to note that PCR-detected RNA fragments do not represent live virus; high RNA load does not necessarily equate to high infectivity. Although studies have shown a good correlation between peak RNA load and infectious virus titer, particularly in the two days surrounding the RNA detection peak41. There is also an association between viral load and viral viability, as demonstrated in a meta-analysis of SARS-CoV-242. Additionally, the size of the amplicon significantly impacts the sensitivity and efficiency of qPCR assays. Generally, shorter amplicons are preferred because they amplify more efficiently and are less prone to degradation, resulting in more accurate viral load quantification and detection of viral shedding43. However, this also means that qPCR-detected viral RNA shedding may last much longer than the shedding of infectious viral particles. Some studies suggest that defining CDV recovery based on the time frame of peak viral load shedding can significantly reduce isolation time for dogs44. However, for endangered wildlife conservation, this decision should be made with more caution. Taking the endpoint of viral RNA shedding detection as a reference for the end of isolation is clearly a more convenient and reliable choice for monitoring.
Our study suggests that vaccinated dogs have the potential to transmit infectious viral particles through shedding, thereby endanger susceptible animals. Furthermore, evidence of viral shedding was noted in these subsequently infected animals. This highlights the concern that free-roaming vaccinated dogs could potentially posing a risk to nearby giant pandas. More in-depth investigations and monitoring of the potential role of viral load during shedding and the survival time of the virus shed in the environment are needed. The purpose of this study is not to discourage vaccination, but to highlight its importance. Results of the study showed that dogs with high initial antibody levels displayed a significantly shorter duration and lower level of viral shedding compared with dogs with low antibody levels. Pre-existing antibodies can reduce the replication and shedding of vaccine viruses by neutralizing them45. After vaccination, the infectivity of individuals and the viral load of transmissible virus during shedding can be significantly reduced46. Our study showed that the presence of high levels of neutralizing antibodies led to a reduced scale of viral shedding, which was also observed in two studies on the viral shedding from COVID-19 patients47,48. This demonstrates the necessity of regular vaccination for domestic dogs to maintain high antibody levels and reduce the risk of viral shedding. Theoretically, dogs with elevated levels of CDV antibodies, even in the absence of a specific titer, exhibit protective immunity and pose a reduced threat to the wild giant panda population and its habitat, as evidenced by the reduction in viral shedding. This conclusion is supported by previous studies that have established the presence of CDV antibodies as a hallmark of protective immunity, regardless of their titer levels49,50. Considering the high prevalence of canine distemper in China51 and the low vaccination rate in domestic dogs around nature reserves, it is imperative to vaccinate dogs with core vaccines once a year to maintain high antibody levels. Furthermore, our study found that domestic dogs with high levels of antibodies, as measured by the duration and amount of viral shedding, posed a lower risk to coexisting animals. Despite the high cost of antibody testing, evidence is scarce from large-scale studies on antibody levels in dogs around nature reserves in China. Therefore, from the standpoint of protecting giant pandas, it is advisable to regularly vaccinate dogs in the vicinity of giant panda habitats.
The recombinant vaccines against CDV, like canarypox virus-based vaccines, typically do not replicate in non-avian cells, minimizing the risk of viral shedding, making them potentially suitable for the protection of endangered wildlife. However, limitations in cost and availability prevent us from widely applying the recombinant CDV vaccines to dogs around the reserves to establish an immunization barrier. These recombinant vaccines have been proven safe and effective in various species, including minks and red pandas52,53. In two studies on giant pandas, involving 5 cubs and 2 adults vaccinated with the canarypox-vectored canine distemper virus, no significant adverse reactions were observed, indicating promising safety results13,54. However, the seroconversion effectiveness was not satisfactory. There is a need for large-scale vaccine studies in giant pandas, including both juvenile and adult individuals, to obtain more conclusive evidence.
The origin of the high initial antibody levels observed in the studied dogs, who were previously free-ranging and un-sheltered, is not definitively known and could have resulted from either natural infection or prior vaccination. Furthermore, the long-term monitoring of viral shedding patterns in these dogs after repeated vaccinations and natural infections calls for further collaborative investigations. Additionally, it is important to investigate the impact of live vaccines used in domestic dog vaccinations near wildlife habitats. These studies should focus on virus shedding and its potential effects on wildlife epidemics. Currently, research in this area, particularly within giant panda national parks, is limited, highlighting the need for more comprehensive studies. We recommend integrating interdisciplinary approaches, including zoology, ecology, vaccinology, and wildlife management, to develop a thorough understanding of disease prevention and control. This is especially crucial for highly infectious and deadly diseases like canine distemper, which pose significant risks to wildlife populations.
Our findings support the notion that proper management of domestic dog vaccination in the proximity of nature reserves can help establish protective barriers against CDV. However, post-vaccination management must be given equal consideration to ensure that the risk of exposure to vaccine strains among wild giant pandas is not heightened.Restricting the activities of vaccinated dogs for at least three weeks post-vaccination is advisable due to ongoing viral shedding. This study highlights the importance of implementing a comprehensive post-vaccination management strategy for dogs residing in giant panda habitats.
Materials and methods
Animals groups
This study involved a total of 69 healthy dogs raised at observation sites in mountainous areas near the Sichuan Fengtongzhai National Nature Reserve in Ya’an. The dogs, which ranged in age from 9 months to 6 years and represented several different breeds, had their age, sex, and body weight recorded. These dogs had no history of receiving canine core vaccines. The animal study, including sample collection, was reviewed and approved by the Animal Care and Use Committee of China Agricultural University (Approval Number: AW01404202-2-1).
We randomly divided the dogs into five groups, as described in Appendix A. Group A, B, and C comprised 18 dogs each and were injected with one of three commercially available live attenuated vaccines Vanguard Plus 5 (Zoetis Inc., USA), NOBIVAC DHPPI (INTERVET, Netherlands), and EURICAN DHPPI2-L (MERIAL, France). Based on the levels of CDV antibodies detected before vaccination, these three groups were further divided into high and low initial antibody level subgroups. The low and high initial antibody level was defined based on the median initial CDV antibody detected in 54 vaccinated dogs on Day 0.
Dogs in group A, B and C were single housed in kennels (2.0 m L by 1.0 m W by 1.5 m H). Nine dogs in group D were not vaccinated and housed separately in three kennels (4.0 m L by 2.0 m W by 1.5 m H), with each kennel accommodating one dog with low antibody level from either group A, B, or C. Specifically, the three dogs in group Da (b and c) were housed with a dog from group AL (BL and CL). Group E comprised six dogs that were not vaccinated and single housed as the control group. The 69 dogs lived in a closed environment, with each group housed in its kennel to prevent interaction between the groups. The facility offered rigorously regulated environmental parameters, including controlled light cycle (12-hour light / 12-hour dark), temperature (21 to 22℃), and humidity (55 to 60%) conditions.
Detection of CDV antibody (IgG) by ELISA
The day of vaccination was designated as Day 0. Blood samples (5 mL) were collected from the cephalic vein on Day 0 before vaccination and Day 21. CDV antibodies were detected by indirect ELISA using a canine distemper virus antibody (IgG) detection kit (Ingenasa, Spain) following the manufacturer’s instructions. The average OD value of each sample was obtained by measuring at 450 nm in a spectrophotometer (Berten Instrument Co., Ltd). The control samples were used to calculate cut-off values for the assays and to determine assay specificity. We calculated the IgG concentration using a 4-parameter logistic (4-PL) model, with the equation y=(A-B)/[1+(x/C)^D] + B where y is the response, A and B are the response at the high and low asymptotes of the curve, x is the analyte concentration, C is the inflection point on the calibration curve (IC50), and D is a slope factor.
Virus detection by qRT-PCR
We collected ocular and nasal swabs, urine, and feces samples daily from Day 0 to Day 21. Total RNA was extracted using a DNA/RNA kit for Virus Detection (Aidlab Biotechnologies Co., Ltd). We synthesized cDNA using an EasyScript One-Step RT-PCR SuperMix Kit (TransGen Biotech Co., Ltd), which was carried out at 42 °C for 30 min (for PCR) or 15 min (for qPCR), followed by a denaturation step at 85 °C for 5 s.
Based on previous study43, we designed primers and a TaqMan probe using Primer 6.0 software (as described in Appendix B). They were synthesized by Sangon Biotech (Shanghai), based on the sequence of CDV Onderstepoort strain (GenBank: AF305419). The TaqMan-based qRT-PCR technique was used to monitor viral shedding, where the TaqMan probe was labeled with 6-carboxyfluorescein (FAM) at the 5’ end and 6-carboxy tetramethylrhodamine (TAMRA) at the 3’ end. To create a standard curve, the RNA from the CDV Onderstepoort strain was prepared and used to construct a standard RNA using the primers S-F and S-R. The RT-PCR product was then cloned into a PTOPO-TA vector and transcribed by Sangon Biotech (Shanghai). The N gene transcript was confirmed through DNA sequencing and quantitated using spectrophotometry on a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific Inc.). The standard RNA was diluted ranging from 8 × 102 copies/µL to 8 × 108 copies/µL (10-fold serial dilutions). Plasmid standards were then used to generate a standard curve for the calculation of viral concentrations in the samples. The standard equation was calculated based on this standard curve. Real-time PCR conditions consisted of an initial denaturation step of 15 min at 95 °C, followed by 37 cycles of 8 seconds at 95 °C and 32 s at 60 °C. The viral RNA copies in the samples were calculated according to the standard curve equation.
Phylogenetic analysis
To verify the origin of virus shedding in group D, we sequenced the hemagglutinin (H) gene from positive ocular and nasal secretion samples collected throughout the entire period of positive detection in the nine group D dogs, along with isolates from three vaccines. Full-length H gene sequences, representative of these viral strains, were obtained, amplified using H-F and H-R primers (Appendix B). A phylogenetic tree was generated based on the H gene. We compared these isolates with 51 global reference strains from GenBank (Appendix D). Sequence alignment was conducted using the ClustalW method in MEGA 10 software, and the phylogenetic tree was constructed using the Maximum Likelihood method and the Tamura-Nei model, with 1000 bootstrap replicates. The reaction condition of RT-PCR was set as follows: initial denaturation at 94 °C for 3 min, followed by 37 cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 60 s. We submitted the H gene sequences of nine isolates from group D and isolates from three vaccines to GenBank (GenBank accession numbers: OQ282897-OQ282908).
Habitat area overlap analysis
We tracked 20 local domestic dogs from villages around the Foping Nature Reserve (FPNR) and Fengtongzhai Nature Reserve (FTNR), which were free-ranging and frequented the core areas of the reserves (FPNR, n = 8; FTNR, n = 12; Appendix E). The dogs were fitted with GPS collars (Lotek GPS3300SL, Lotek Wireless Inc., Newmarket, Ontario, Canada), and their locations were recorded every hour from 06:00 to 18:00, with 12 position points recorded daily. The recorded GPS data also included time, date, longitude, latitude, elevation, satellite number, and the position precision of the attenuation factor. The mobile loci of 6 giant pandas in the FTNR were obtained through observational methods, while those in the FPNR were obtained through GPS collars on 6 wild pandas from 2012 to 2015. Geographic analyses were conducted using ArcGIS v10.5 (Environmental Systems Research Institute Inc., Redlands, USA). We used the Fixed Kernel Estimation (FKE) method and Minimum Convex Polygon (MCP) analysis to estimate the core areas and overall overlap of habitat areas of giant pandas and domestic dogs.
Statistical analysis
Data analysis was performed using the software SPSS 25.0. We analyzed the changes in antibody levels after vaccination using a one-sample K-S test and one-way analysis of variance (ANOVA). The standard curve of ELISA was determined using a four-parameter logistic model, and the standard curve of qRT-PCR was determined using linear regression analysis (least-squares method). To study the viral shedding after vaccination, one-way ANOVA was performed and the results of viral shedding detected in the vaccine groups were displayed by GraphPad Prism 8.0. Based on previous studies55,56, GPS points were only selected if the 3D positioning and positional dilution of precision values were < 10. We used the geographic information system software ArcGIS v10.5 (Environmental Systems Research Institute Inc., Redlands, USA) to generate the maps of the study area.
Data availability
The datasets and accession number presented in this study can be found in the article/supplementary material.
References
-
Hu, J. Research history and taxonomy of giant panda. Bull. Biology, 1–4 (1990).
-
State Forestry Administration. Release of the fourth national survey report on giant panda in China, (2015). http://www.forestry.gov.cn/main/4462/20150303/743596.html
-
Wei, F. et al. Progress in the ecology and conservation of giant pandas. Conserv. Biol. 29, 1497–1507. https://doi.org/10.1111/cobi.12582 (2015).
-
Wang, C. et al. Isolation and identification of rotavirus from giant panda cubs. Acta Theriol. Sinica. 87–91. https://doi.org/10.16829/j.slxb.2008.01.014 (2008).
-
Qin, Q. et al. Serosurvey of selected viruses in captive giant pandas (Ailuropoda melanoleuca) in China. Vet. Microbiol. 142, 199–204. https://doi.org/10.1016/j.vetmic.2009.09.062 (2010).
-
Rendon-Marin, S., da Fontoura Budaszewski, R., Canal, C. & Ruiz-Saenz, J. Tropism and molecular pathogenesis of canine distemper virus. Virol. J. 16 https://doi.org/10.1186/s12985-019-1136-6 (2019).
-
Mainka, S., Qiu, X., He, T. & Appel, M. Serologic survey of giant pandas (Ailuropoda melanoleuca), and domestic dogs and cats in the Wolong Reserve, China. J. Wildl. Dis. 30, 86–89. https://doi.org/10.7589/0090-3558-30.1.86 (1994).
-
Li, J. et al. Gene sequence analysis diagnosis of Giant pandas infected by Canine Distemper Virus. Chin. J. Veterinary Sci. 448–450. https://doi.org/10.16303/j.cnki.1005-4545.1999.05.010 (1999).
-
Feng, N. et al. Fatal canine distemper virus infection of giant pandas in China. Sci. Rep. 6, 27518. https://doi.org/10.1038/srep27518 (2016).
-
Jin, Y. et al. Canine distemper viral infection threatens the giant panda population in China. Oncotarget. 8, 113910–113919. https://doi.org/10.18632/oncotarget.23042 (2017).
-
Sun, J. et al. Changes in the MicroRNA Profile of the Giant Panda after Canine Distemper Vaccination and the Integrated Analysis of MicroRNA-Messenger RNA. DNA Cell Biol. 40, 595–605. https://doi.org/10.1089/dna.2020.5942 (2021).
-
Wang, C., Yang, S., Wu, K., Gao, Y. & Zhang, Z. Serological evaluation of the efficacy of the multivalent canine distemper attenuated live vaccines on giant pandas (Ailuropoda melanoleuca). Acta Theriol. Sinica. 212–216. https://doi.org/10.16829/j.slxb.2008.02.015 (2008).
-
Geng, Y. et al. First demonstration of giant panda’s immune response to canine distemper vaccine. Dev. Comp. Immunol. 102, 103489. https://doi.org/10.1016/j.dci.2019.103489 (2020).
-
Jin, Y., Liu, Q., Sun, M., Qiao, Y. & Qiao, M. Genomic characterization of the newly emerged Canine Distemper Virus in Giant Panda. Scientia Agricultura Sinica. 48, 1445–1452 (2015).
-
Cleaveland, S., Kaare, M., Knobel, D. & Laurenson, M. Canine vaccination–providing broader benefits for disease control. Vet. Microbiol. 117, 43–50. https://doi.org/10.1016/j.vetmic.2006.04.009 (2006).
-
Cleaveland, S. et al. The Conservation Relevance of Epidemiological Research into Carnivore viral diseases in the Serengeti. Conserv. Biol. 21, 612–622. https://doi.org/10.1111/j.1523-1739.2007.00701.x (2007).
-
Kotani, T., Jyo, M., Odagiri, Y., Sakakibara, Y. & Horiuchi, T. Canine distemper virus infection in lesser pandas (Ailurus fulgens). Nihon Juigaku Zasshi. Japanese J. Veterinary Sci. 51, 1263–1266. https://doi.org/10.1292/jvms1939.51.1263 (1989).
-
Perpiñán, D., Ramis, A., Tomás, A., Carpintero, E. & Bargalló, F. Outbreak of canine distemper in domestic ferrets (Mustela putorius furo). Vet. Rec. 163, 246–250. https://doi.org/10.1136/vr.163.8.246 (2008).
-
Appel, M. et al. Canine distemper epizootic in lions, tigers, and leopards in North America. J. Vet. Diagn. Invest. 6, 277–288. https://doi.org/10.1177/104063879400600301 (1994).
-
Osterhaus, A. et al. Canine distemper virus in seals. Nature. 335, 403–404. https://doi.org/10.1038/335403a0 (1988).
-
Sakai, K. et al. Lethal canine distemper virus outbreak in cynomolgus monkeys in Japan in 2008. J. Virol. 87, 1105–1114. https://doi.org/10.1128/jvi.02419-12 (2013).
-
Loots, A., Mitchell, E., Dalton, D., Kotzé, A. & Venter, E. Advances in canine distemper virus pathogenesis research: a wildlife perspective. J. Gen. Virol. 98, 311–321. https://doi.org/10.1099/jgv.0.000666 (2017).
-
Roelke-Parker, M. et al. A canine distemper virus epidemic in Serengeti lions (Panthera leo). Nature. 379, 441–445. https://doi.org/10.1038/379441a0 (1996).
-
Williams, E., Thorne, E., Appel, M. & Belitsky, D. Canine distemper in black-footed ferrets (Mustela nigripes) from Wyoming. J. Wildl. Dis. 24, 385–398. https://doi.org/10.7589/0090-3558-24.3.385 (1988).
-
Millán, J. et al. Serosurvey of dogs for human, livestock, and wildlife pathogens, Uganda. Emerg. Infect. Dis. 19, 680–682. https://doi.org/10.3201/eid1904.121143 (2013).
-
Bronson, E., Emmons, L. H., Murray, S., Dubovi, E. J. & Deem, S. L. Serosurvey of pathogens in domestic dogs on the border of Noël Kempff Mercado National Park, Bolivia. J. Zoo Wildl. Med. 39, 28–36. https://doi.org/10.1638/2006-0046.1 (2008).
-
Shen, T., Welburn, S. C., Sun, L. & Yang, G. J. Progress towards dog-mediated rabies elimination in PR China: a scoping review. Infect. Dis. Poverty. 12 https://doi.org/10.1186/s40249-023-01082-3 (2023).
-
Liu, Z., Liu, M., Tao, X. & Zhu, W. Epidemic characteristics of human rabies – China, 2016–2020. China CDC Wkly. 3, 819–821. https://doi.org/10.46234/ccdcw2021.203 (2021).
-
Yan, X. et al. Dogs and Disease threats to Giant pandas in China. J. Wildl. Manag. 84, 268–276. https://doi.org/10.1002/jwmg.21786 (2020).
-
Fèvre, E., Bronsvoort, B., Hamilton, K. & Cleaveland, S. Animal movements and the spread of infectious diseases. Trends Microbiol. 14, 125–131. https://doi.org/10.1016/j.tim.2006.01.004 (2006).
-
Acosta-Jamett, G. et al. Urban domestic dog populations as a source of canine distemper virus for wild carnivores in the Coquimbo region of Chile. Vet. Microbiol. 152, 247–257. https://doi.org/10.1016/j.vetmic.2011.05.008 (2011).
-
Wilkes, R. P. Canine Distemper Virus in Endangered Species: Species Jump, Clinical Variations, and Vaccination. Pathogens 12, doi: (2022). https://doi.org/10.3390/pathogens12010057
-
Kapil, S. & Yeary, T. J. Canine distemper spillover in domestic dogs from urban wildlife. Veterinary Clin. North. America: Small Anim. Pract. 41, 1069–1086. https://doi.org/10.1016/j.cvsm.2011.08.005 (2011).
-
Bush, M., Montali, R. J., Brownstein, D., James, A. E. Jr. & Appel, M. J. Vaccine-induced canine distemper in a lesser panda. J. Am. Vet. Med. Assoc. 169, 959–960 (1976).
-
Carpenter, J., Appel, M., Erickson, R. & Novilla, M. Fatal vaccine-induced canine distemper virus infection in black-footed ferrets. J. Am. Vet. Med. Assoc. 169, 961–964 (1976).
-
Sutherland-Smith, M. et al. Vaccine-induced canine distemper in European mink, Mustela lutreola. J. zoo Wildl. Medicine: Official Publication Am. Association Zoo Veterinarians. 28, 312–318 (1997).
-
Sattler, U. et al. Identification of amino acid substitutions with compensational effects in the attachment protein of canine distemper virus. J. Virol. 88, 8057–8064. https://doi.org/10.1128/jvi.00454-14 (2014).
-
Fischer, C. D. B. et al. Phylogenetic analysis of canine distemper virus in South America clade 1 reveals unique molecular signatures of the local epidemic. Infect. Genet. Evol. 41, 135–141. https://doi.org/10.1016/j.meegid.2016.03.029 (2016).
-
Demeter, Z., Palade, E. A., Hornyák, A. & Rusvai, M. Controversial results of the genetic analysis of a canine distemper vaccine strain. Vet. Microbiol. 142, 420–426. https://doi.org/10.1016/j.vetmic.2009.10.017 (2010).
-
Tamukai, K. et al. Molecular evidence for vaccine-induced canine distemper virus and canine adenovirus 2 coinfection in a fennec fox. J. Vet. Diagn. Invest. 32, 598–603. https://doi.org/10.1177/1040638720934809 (2020).
-
Sehata, G. et al. Use of quantitative real-time RT-PCR to investigate the correlation between viremia and viral shedding of canine distemper virus, and infection outcomes in experimentally infected dogs. J. Vet. Med. Sci. 77, 851–855. https://doi.org/10.1292/jvms.14-0066 (2015).
-
Cevik, M. et al. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2, e13–e22. https://doi.org/10.1016/s2666-5247(20)30172-5 (2021).
-
Elia, G. et al. Detection of canine distemper virus in dogs by real-time RT-PCR. J. Virol. Methods. 136, 171–176. https://doi.org/10.1016/j.jviromet.2006.05.004 (2006).
-
Allen, C., Ellis, A., Liang, R., Lim, A. & Newbury, S. Prolonged persistence of canine distemper virus RNA, and virus isolation in naturally infected shelter dogs. PLOS ONE. 18, e0280186. https://doi.org/10.1371/journal.pone.0280186 (2023).
-
Mok, D. Z. L. & Chan, K. R. The Effects of Pre-Existing Antibodies on Live-Attenuated Viral Vaccines. Viruses 12, doi: (2020). https://doi.org/10.3390/v12050520
-
Puhach, O., Meyer, B. & Eckerle, I. SARS-CoV-2 viral load and shedding kinetics. Nat. Rev. Microbiol. 21, 147–161. https://doi.org/10.1038/s41579-022-00822-w (2023).
-
Yin, Y. et al. Impact of antibody-level on viral shedding in B.1.617.2 (Delta) variant-infected patients analyzed using a joint model of longitudinal and time-to-event data. Math. Biosci. Eng. 20, 8875–8891. https://doi.org/10.3934/mbe.2023390 (2023).
-
Lee, Y. L. et al. Dynamics of anti-SARS-Cov-2 IgM and IgG antibodies among COVID-19 patients. J. Infect. 81, e55–e58. https://doi.org/10.1016/j.jinf.2020.04.019 (2020).
-
Schultz, R. D., Thiel, B., Mukhtar, E., Sharp, P. & Larson, L. J. Age and long-term protective immunity in dogs and cats. J. Comp. Pathol. 142 (Suppl 1), 102–108. https://doi.org/10.1016/j.jcpa.2009.10.009 (2010).
-
Belsare, A. V. & Gompper, M. E. To vaccinate or not to vaccinate: lessons learned from an experimental mass vaccination of free-ranging dog populations. Anim. Conserv. 18 (2015).
-
Qiao, Y. et al. The spatial distribution of prevalence of canine distemper in China (2004–2014). Chin. J. Veterinary Med. 51, 59–60 (2015).
-
Ramsay, E. C., Georoff, T. A., Burrell, C., Anis, E. & Wilkes, R. P. Red pandas’ (Ailurus fulgens) serological response to canarypox-vectored canine distemper vaccines. J. Zoo Wildl. Med. 50, 478–481. https://doi.org/10.1638/2018-0160 (2019).
-
Gong, Y. et al. A highly efficient recombinant canarypox virus-based vaccine against canine distemper virus constructed using the CRISPR/Cas9 gene editing method. Vet. Microbiol. 251, 108920. https://doi.org/10.1016/j.vetmic.2020.108920 (2020).
-
Bronson, E., Deem, S. L., Sanchez, C. & Murray, S. Serologic response to a canarypox-vectored canine distemper virus vaccine in the giant panda (Ailuropoda melanoleuca). J. Zoo Wildl. Med. 38, 363–366. https://doi.org/10.1638/1042-7260 (2007).
-
D’eon, R. G., Serrouya, R., Smith, G. C. & Kochanny, C. O. GPS radiotelemetry error and bias in mountainous terrain. Wildl. Soc. Bull. 30, 430–439 (2002).
-
Lewis, J. S., Rachlow, J. L., Garton, E. O. & Vierling, L. A. Effects of habitat on GPS collar performance: using data screening to reduce location error. J. Appl. Ecol. 44, 663–671. https://doi.org/10.1111/j.1365-2664.2007.01286.x (2007).
Acknowledgements
We are grateful to W. Wei for his invaluable contribution to GPS tracking data and support in the preparation of this manuscript. We are grateful to three anonymous reviewers for useful comments on an earlier version of this paper. We thank those who helped recruit dogs and collect samples in this study, most notably W. Feng, W. Gang, S. Xianpeng, D. Jiaqiang, and S. Jie.
Author information
Authors and Affiliations
Contributions
ZF, HS, GL, YJ: conceptualization. ZF, ZL, QW, ST: sample collections. ZF, QW, YW: data analysis. ZF, HS: original draft preparation. ZF, HS, ZL, YW, GL, YJ: review and editing. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical Statement
The animal study including sample collection was reviewed and approved by the Animal Care and Use Committee of China Agricultural University (Approval Number: AW01404202-2-1). All experiments were performed in accordance with relevant guidelines and regulations. The study complied with ARRIVE guidelines.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Fang, Z., Shi, H., Wang, Q. et al. Risk of canine distemper virus vaccination of domestic dogs in giant panda habitat to giant pandas.
Sci Rep 14, 29761 (2024). https://doi.org/10.1038/s41598-024-79806-0
-
Received:
-
Accepted:
-
Published:
-
DOI: https://doi.org/10.1038/s41598-024-79806-0
Keywords
This post was originally published on this site be sure to check out more of their content.